JET2DNA
A tool combining sequence conservation, interface residue propensities and geometry of the protein surface for predicting DNA-binding sites on protein structures.
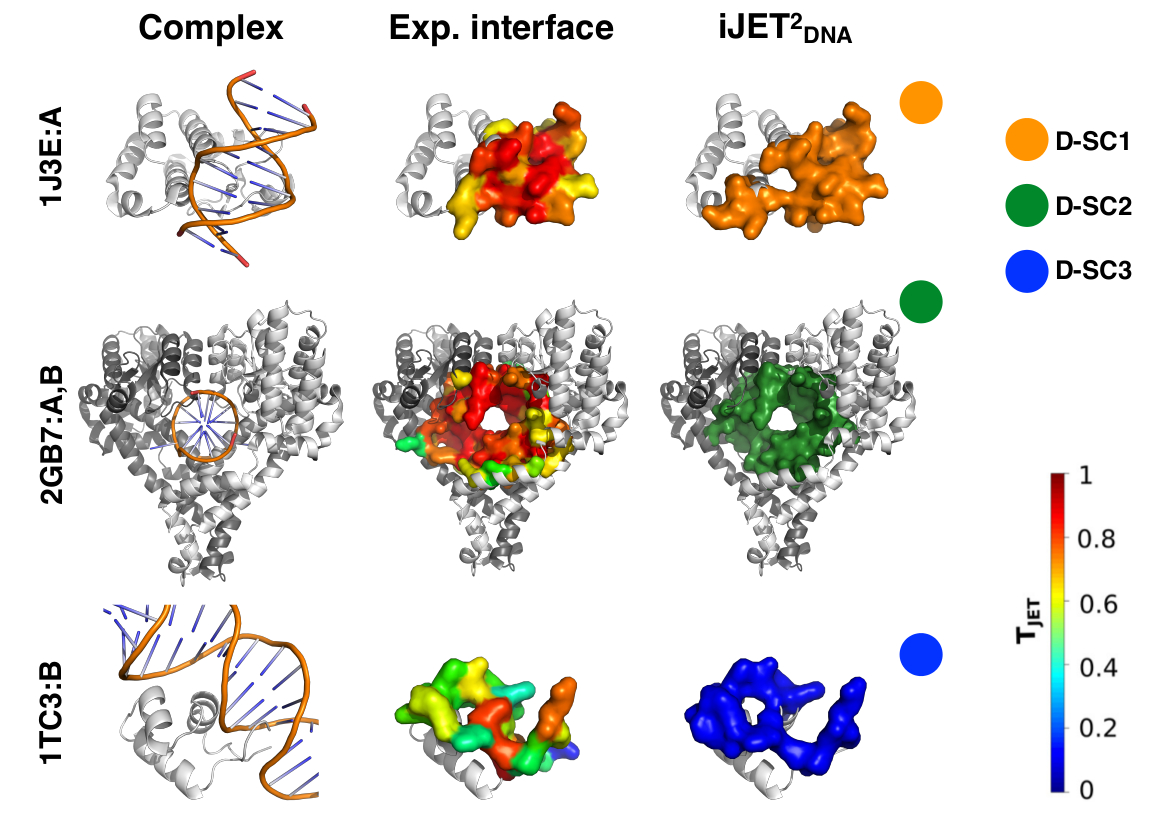 JET2DNA is a new tool for predicting patches of interaction with DNA on protein structures. It combines only three sequence- and structure-based descriptors: sequence evolutionary conservation, interface residue propensities and geometry of the protein surface. These descriptors are combined in three different scoring strategies aimed at predicting a wide range of protein-DNA interfaces. In particular, JET2DNA is able to unravel multiple binding sites on a same protein surface and its predictions are robust to conformational changes. Beyond its predictive power, JET2DNA provides a unique way to understand the origins and properties of the detected interaction sites and interpret those in light of their functions. It can be used in a fully automated mode, where the most appropriate scoring strategy is determined depending on the query protein. Alternatively, the user can modify all parameters depending on the biological question asked. JET2DNA is based on the Joint Evolutionary Trees (JET) method and it is an adaptation of JET2, predicting protein-protein interfaces.
JET2DNA is a new tool for predicting patches of interaction with DNA on protein structures. It combines only three sequence- and structure-based descriptors: sequence evolutionary conservation, interface residue propensities and geometry of the protein surface. These descriptors are combined in three different scoring strategies aimed at predicting a wide range of protein-DNA interfaces. In particular, JET2DNA is able to unravel multiple binding sites on a same protein surface and its predictions are robust to conformational changes. Beyond its predictive power, JET2DNA provides a unique way to understand the origins and properties of the detected interaction sites and interpret those in light of their functions. It can be used in a fully automated mode, where the most appropriate scoring strategy is determined depending on the query protein. Alternatively, the user can modify all parameters depending on the biological question asked. JET2DNA is based on the Joint Evolutionary Trees (JET) method and it is an adaptation of JET2, predicting protein-protein interfaces.
Download
The JET2DNA package is available here.
The HR-PDNA187 and HOLO-APO82 benchmarks of protein-DNA complexes used to test JET2DNA are available here.
System Requirements
- UNIX platform (Linux/OSX).
The program JET2DNA requires some external tools that should be installed:
- java6 or higher. If you have a version of java higher than java6, you could need to recompile the JET2DNA package. To compile it, place yourself in the JET2DNA directory and execute the command make.
- ClustalW v2.1, a tool for performing multiple alignment of nucleic acid and protein sequences (Thompson JD, Higgins DG, Gibson TJ. (1994), Nucleic Acids Research, 22:4673-4680). It is run with either Blosum62, Gonnet or HSDM matrix by automatic selection.
- Naccess v2.1.1, a program that calculates the accessible area of a molecule from a PDB (Protein Data Bank) format file (Hubbard S. J., Thornton J. M. (1993), University College London).
- PSI-BLAST from BLAST+ Toolkit (v2.2.27 or more recent) (Altschul SF, Gish W, Miller W, Myers EW, Lipman D. (1990) J Mol Biol., 215, 403–410). The program can be called on the sever or locally. To reach similar outputs on the server and on a local machine, a local call to PSI-BLAST is coded in JET2DNA, that uses the option -t 2 setting the composition-based score adjustment method conditioned on sequence properties. To run the program locally, the appropriate BLAST databases need to be downloaded.
Installation
- Unzip the JET2DNA.zip in the directory of your choice.
- Set up JET2DNA home directory in your .bashrc or .cshrc file:
export JET2DNA_PATH = path_of_JET2DNA_HOME_directory (bash syntax)
setenv JET2DNA_PATH path_of_JET2DNA_HOME_directory (csh syntax)Running JET2DNA
The command line to execute JET2DNA is:
java -cp $JET2_PATH:$JET2_PATH/jet/extLibs/vecmath.jar jet.JETJET2DNA offers a number of functionalities whose choice is controlled by the option -p:
- A Compute the accessibility surface areas of the atoms and residues using Naccess.
- V Compute the circular variances of the residues.
- J Launch Joint Evolutionary Trees analysis to evaluate residues conservation levels.
- C Run the clustering algorithm to define binding patches.
- G Insert JET2DNA specified values in the B-factors column of a PDB file that will be located in the output directory.
JET2DNA proposes different clustering strategies whose choice is controlled by the option -a:
- 0 The most appropriate strategy is automatically determined by JET2 .
- 1 D-SC1: cluster seed and extension are detected by combining conservation levels and interface propensities; an outer layer is added based on a combination of conservation levels and local circular variance.
- 2 D-SC2: cluster seed and extension are detected by combining conservation levels and global circular variance; an outer layer is added based on a combinantion of conservation levels and interface propensities.
- 3 D-SC3: cluster seed, extension and outer layer are detected from a combination of interface propensities and local circular variance.
Example of a JET2DNA analysis:
- Copy the default.conf configuration file from your $JET2_PATH directory in the directory of your choice (ex: your output directory) and modify the locations of the softwares and the BLAST database and the parameters values you wish. The default.conf configuration file will be modified during JET2DNA calculations and thus it is important to have one default.conf configuration file for each process running in parallel.
- Launch JET2DNA:
java -cp $JET2DNA_PATH:$JET2DNA_PATH/jet/extLibs/vecmath.jar jet.JET -c <default.conf> -i <PDB_ID.pdb> -o <output_directory> -p AVJC -r local -a 3 -d chain
The arguments given are:
- path to the configuration file to be used (-c)
- path to the input PDB file (-i)
- the directory where output files should be stored (-o)
- the type(s) of analysis to run (-p)
- the mode for PSI-BLAST call (-r), either on the server (server) or locally (local). If the PSI-BLAST alignment file was already computed, this option can be setted on input and the directory containing the PSI-BLAST alignement file can be specified by the option -b (see below)
- the clustering strategy (-a [0=automatic_choice || 1=D-SC1 || 2=D-SC2 || 3=D-SC3])
- the way the input PDB file should be treated (-d), either as a complex (complex) or by considering each chain individually (chain)
The retrieval of homologous sequences from BLAST database may take some time. It is possible to do it once for all, and then run only JET2DNA and clustering analyses by specifying the -r input option and the location of the PSI-BLAST alignment file (-b):
java -cp $JET2DNA_PATH:$JET2DNA_PATH/jet/extLibs/vecmath.jar jet.JET
-c <default.conf> -i <PDB_ID.pdb> -o <output_directory> -p AVJC -r input -b <dir_psiblast_files> -a 3 -d chainAn iterative version of JET2DNA (iJET2DNA) is also proposed for large-scale predictions. iJET2DNA provides a list of consensus residues belonging to predicted interaction patches and enables to explore the set of potentially interacting residues by varying the consensus threshold. Typically consensus residues detected in at least 2 iterations are considered as robust prediction. iJET2DNA is called using the -n option:
java -cp $JET2DNA_PATH:$JET2DNA_PATH/jet/extLibs/vecmath.jar jet.JET
-c <default.conf> -i <PDB_ID.pdb> -o <output_directory> -p AVJC -r local -a 3 -d chain -n 10To insert JET2DNA results (ex: "clustersOccur" values stored in the pdbID_jet.res file) in the B-factors column of a PDB file, that will be located in the output directory (ex: pdbID_clustersOccur.pdb), use the -p G analysis and the -g option to specify the values to store in the B-factor column:
java -cp $JET2DNA_PATH:$JET2DNA_PATH/jet/extLibs/vecmath.jar jet.JET
-c <default.conf> -i <PDB_ID.pdb> -o <output_directory> -p G -g clustersOccur,traceMax,pc,cv,cvlocalLicence
The JET2DNA package has been developed under the CeCILL licence (see LICENCE).
Contact
For questions, comments or suggestions feel free to contact Flavia Corsi, Elodie Laine or Alessandra Carbone.



