
Elodie Laine
Full Professor in Computational Biology and Artificial Intelligence. Laboratory of Computational and Quantitative Biology, IBPS, Sorbonne Université.
- Head of the PROMISE project, 2023-2028, Consolidator Grant from the European Research Council.
- Junior member of the institut Universitaire de France, 2023-2028.
Accreditation to supervise research (2020) from Sorbonne Université. Young Researcher grant (2018-2021, MASSIV project) from the French National Research Agency. Postdoc (2009-2012) at the Laboratory of Biology and Applied Pharmacology, Ecole Normale Supérieure de Cachan. PhD in Life Sciences (2006-2009) from Sorbonne Université, Structural Bioinformatics Unit, Institut Pasteur. See my resume.
Digest
In a nutshell
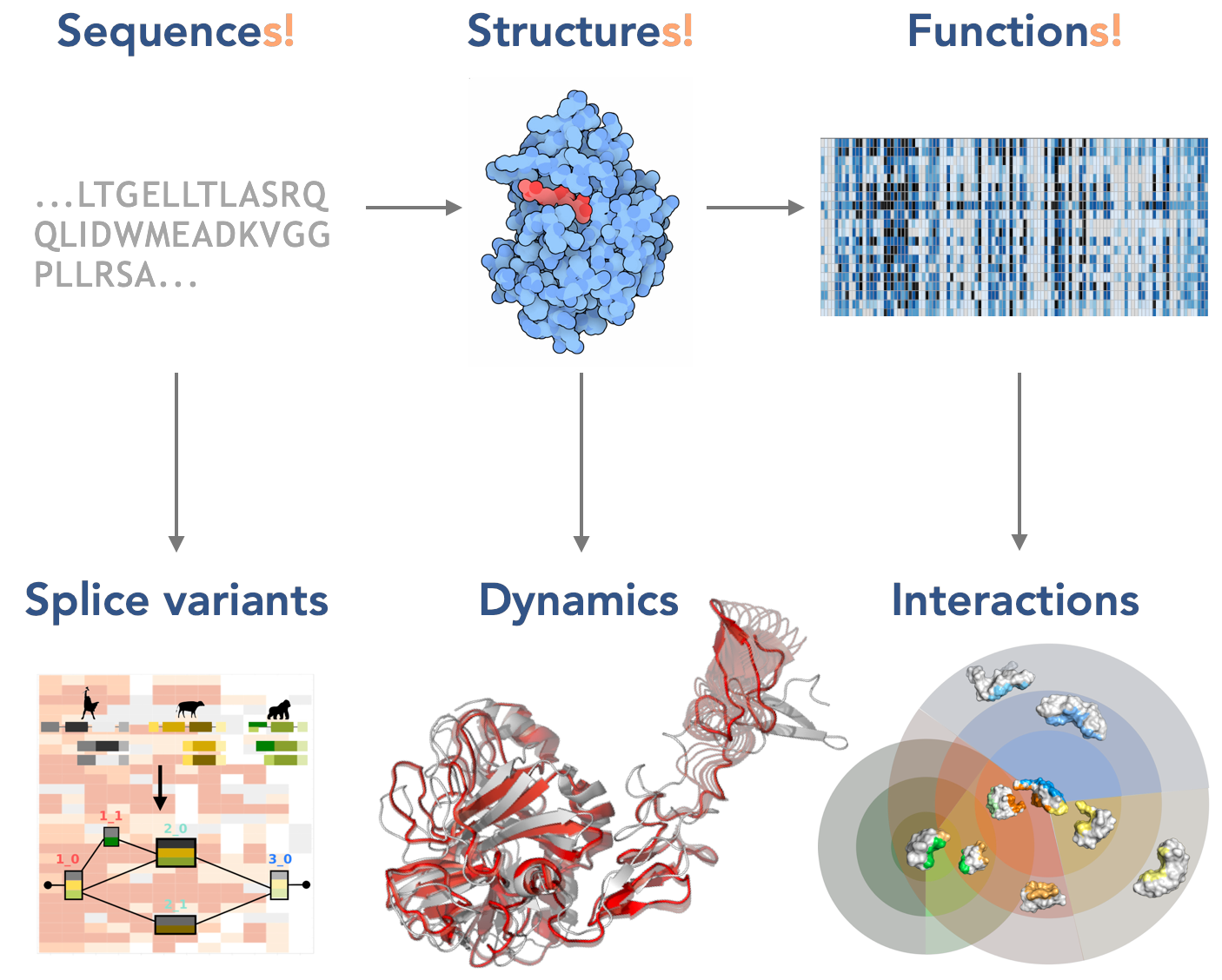
How do proteins fold, move, and associate? How did they evolve? What are the mechanisms responsible for their misfunction? How can computational methods guide biological intervention and the design of better treatments? These questions are at the core of my research. To address them, I have developed a multidisciplinary scientific approach at the interface of biology, computer science, mathematics, and physics.
I have contributed interpretable and scalable methods for assessing protein diversity in evolution, describing protein functional states and motions, predicting protein interfaces and binding partners, and predicting protein mutational landscapes.
I have co-created with Anne Lopes (U. Paris-Saclay, France) an innovative cross-university collaborative course immersing Masters students into the research ecosystem with project-based learning, flipped classrooms, and "coopetition". We published an educational article. I coordinated the implementation of the initiative at the European level with colleagues from the 4EU+ alliance.
Team
Lab and collab

Marina Abakarova
PhD student. Large-scale assessment of the impact of protein sequence variations on ageing using Drosophila melanogaster. Funded by the ANR.
Louis Carrel-Billiard
PhD student. Charting proteoform diversity across 800 million years of evolution. Funded by the ERC.
Valentin Lombard
PhD student. Geometric deep manifold learning combined with Natural Language Processing for protein movies. SCAI doctoral grant.
Julien Nguyen Van
PhD student. Deciphering the complexity of proteoform interactions with evolutionary- and physically-informed protein language models. Funded by the ERC.
Michael Rera
CNRS Researcher at the Interdisciplinary Research Centre (France). Ageing and natural death, Drosophila. Co-advises M. Abakarova.
Sergei Grudinin
CNRS Researcher at Laboratoire Jean Kuntzmann (France). Macromolecular structures and interactions, AI methods for graph learning. Co-advises V. Lombard.
Hugues Richard
Researcher at the Robert Koch Institute (Germany). high-throughput sequencing data, especially transcriptome (RNA-Seq) data and statistical modelling. Co-advised several Post-doc and students, MASSIV project.Alessandra Carbone
Professor at Sorbonne Université (SU), Head of the Laboratory of Computational and Quantitative Biology (LCQB, France). Co-advised 1 Post-doctoral fellow and 4 PhD students on protein interactions and protein mutational landscapes.Béatrice Durand (SU, France) - Wnt signaling, 20K funding from Institute of Biology Paris-Seine, 2023-2024.Mathilde Garcia (SU, France) - Co-translational mechanisms, 6K from SU System Biology Network, 2017.Burkhard Rost (TUM, Germany) - Protein mutational landscapes.
Community efforts - CASP (Zoom), CAPRI, Elixir 3D Bioinfo, ML4NGP, ERGA, LEGO, MASIM.
We are hiring!
We are looking for highly talented and motivated PhDs and Post-doctoral fellows, with backgrounds in applied mathematics, machine learning, algorithms for high-throughput sequencing data, structural bioinformatics and evolutionary biology.
Get in touch!Publications
Full list here

Expert-guided protein Language Models enable accurate and blazingly fast fitness prediction
C. Marquet, J. Schlensok, M. Abakarova, B. Rost, and E. Laine* (2024) Bioinformatics View details »

Explaining Conformational Diversity in Protein Families through Molecular Motions
V. Lombard, S. Grudinin* and E. Laine* (2024) Scientific data View details »

Alignment-based protein mutational landscape prediction: doing more with less
M. Abakarova, C. Marquet, M. Rera, B. Rost, and E. Laine* (2023) Genome Biology and Evolution View details »

Functional mapping of N-terminal residues in the yeast proteome uncovers novel determinants for mitochondrial protein import
S. Nashed, H. El Barbry, M. Benchouaia, A. Dijoux-Maréchal, T. Delaveau, N. Ruiz-Gutierrez, L. Gaulier, D. Tribouillard-Tanvier, G. Chevreux, S. Le Crom, B. Palancade, F. Devaux, E. Laine and M. Garcia* (2023) PLOS Genetics View details »

Mega-scale experimental analysis of protein folding stability in biology and design
K. Tsuboyama, J. Dauparas, J. Chen, E. Laine, Y. Mohseni Behbahani, J. J. Weinstein, N. M. Mangan, S. Ovchinnikov and G. J. Rocklin* (2023) Nature View details »

Building alternative splicing and evolution-aware sequence-structure maps for protein repeats
A. Szatkownik, D.J. Zea, H. Richard* and E. Laine* (2023) J. Struct. Biol. View details »
Software
Try them out
- All
- Diversity
- Interactions
- Mutations
- Motions

ASES [Video]
What does protein diversity look like?
D.J. Zea, H. Richard*, and E. Laine* (2022) Bioinformatics.
ThorAxe [Video]
Which versions of my favourite protein are conserved in evolution?
D.J. Zea, S. Laskina, A. Baudin, H. Richard*, and E. Laine* (2021) Genome Research.
NOLB [Video]
How does my favourite protein move?
S. Grudinin+, E. Laine+, and A. Hoffmann (2020) Biophys. J. A. Hoffmann and S. Grudinin (2017) J. Chem. Th. Comput.
GEMME [Cover]
Is this mutation deleterious?
E. Laine*, Y. Karami and A. Carbone*. (2019) Mol. Biol. Evol.Conferences
Scientific events and courses
LEGO at JOBIM 2024
Half-day meeting on Machine Learning for Genomics and Mini-Symposium shared with StatOmique in Machine Learning and Statistics in Genomics and Metagenomics, June 2024, France.
Deep Learning in Life Sciences
Collaborative course between European Universities, Feb.-June 2024, online. [materials]
LEGO 2023, Machine Learning for Genomics
National working group meeting (90 participants), May 2023, France.
Interplay between AI and mathematical modelling in the post-structural genomics era
International workshop (70 participants), March 2023, France.
AI for Biologists
International summer school (20 students), July 2022, France
New directions of AI in structural biology
International workshop (35 participants), August 2021, France.
Integrative structural modeling in the era of big data and artificial intelligence
Mini-symposium at the French Society for Bioinformatics annual meeting (80 participants), July 2021, online.
Meet-4EU+ Symposium
International symposium mixing education and research (100 participants), January 2021, online.
Noncanonical amino acids (ncAAs): tools for biological and biophysical investigations
International symposium (100 participants), October 2019, France.
Formal methods in the eyes of biological data
Symposium at the French Society for Applied Mathematics meeting (35 participants), June 2017, France.
Evolution of alternative splicing
International symposium at the Society for Molecular Biology and Evolution annual congress, (70 participants), July 2015, Austria
Communications for the general public
Contact
Get in touch
Location:
LCQB, UMR 7238
IBPS, CNRS - Sorbonne Université
Jussieu campus – Building C, 4th floor
7-9, quai Saint-Bernard
75005 Paris, FRANCE
Email:
elodie.laine@sorbonne-universite.fr





























